Alpha-1 antitrypsin deficiency: What protects the one – and not the other?
Researchers at the MPI of Biochemistry have used spatial Deep Visual Proteomics workflow to reveal why some patients with the hereditary disease alpha-1 antitrypsin deficiency remain healthy despite the genetic defect.
As part of an international study, researchers at the Max Planck Institute of Biochemistry used spatial proteomics method called Deep Visual Proteomics to analyze liver tissue from German and Danish patients with Alpha-1 Antitrypsin Deficiency. Their findings explain why two-thirds of patients with the homozygous form of this genetic disorder never develop liver fibrosis, despite carrying the same mutation as those who do. This discovery, published in Nature, provides new opportunities for early detection and prevention strategies for this hereditary condition.
In a nutshell:
- Hereditary disease alpha-1-antitrypsin deficiency: researchers have discovered why some patients remain healthy despite a genetic defect, while others develop severe liver fibrosis
- Deep Visual Proteomics: combines AI imaging with mass spectrometry to reveal distinct protein patterns in liver cells, showing how they get sick and what natural healing mechanisms they activate.
- Hope for clinical application: One possible approach would be the development of a liver fibrosis early warning system based on the early peroxisomal response of liver cells.
In this Video Florian Rosenberger, first author of the publication, explains the workflow of the study.
Alpha-1-antitrypsin is a so-called protease inhibitor, a type of enzyme inhibitor. It is produced in the liver but exerts its effects in the lungs, where it regulates immune cell activity. This regulation is crucial and an overactive immune response can cause serious lung diseases. However, some individuals carry a genetic mutation that causes the alpha-1 protein to fold incorrectly. As a result, too little functional alpha-1 is produced, and insufficient amounts reach the lungs.
The mutation is inherited from one or both parents. Around one in 20 people in Europe carries the heterozygous form of the mutation – inherited from only one parent – and often experience no symptoms or only mild ones. In contrast, the rarer homozygous form, inherited from both parents, affects approximately one in 2,000 individuals and is much more severe. These patients are at a higher risk of developing not only lung disease, such as chronic obstructive pulmonary disease (COPD), but also liver complications including severe fibrosis or even tumors.
One Mutation, Divergent Paths
An international research team led by Prof. Matthias Mann at the Max Planck Institute of Biochemistry in Martinsried, near Munich, has now uncovered important new insights into the homozygous form of the condition. First author Florian Rosenberger, from the Department of Proteomics and Signal Transduction, explains: “In the homozygous form, something striking stands out. It is a monogenetic disease, meaning all patients carry the same mutation – so in theory, disease progression should be uniform. But that’s not what we see. One-third of patients develop severe liver fibrosis, where connective tissue accumulates and impairs liver function. Two-thirds, however, remain healthy. We wanted to understand why that is. What molecular mechanisms protect some patients while others develop disease?”
The team used a technique called Deep Visual Proteomics, developed collaboratively in Martinsried and Copenhagen by the proteomics research groups of Matthias Mann. This method applies advanced proteome analysis to identify disease mechanisms. For the Alpha-1 study, liver tissue samples from patients in Germany and Denmark were analyzed.
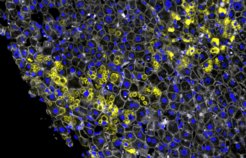
“We examined tissue across the full spectrum of disease stages,” Rosenberger continues. “Even in early stages – when clinical signs had not yet appeared – we could observe how the body in some cases successfully halted disease progression.” For their analysis, the researchers employed a convolutional neural network (CNN) – a form of artificial intelligence originally trained to recognize faces and everyday objects in images.
Subtle Differences with Big Impacts
The CNN also performed impressively on images of human liver tissue. It was able to differentiate between subtle structural variations in the disease, particularly the way alpha-1 protein aggregates in liver cells (hepatocytes). These aggregates are a hallmark of disease onset. “Our CNN could detect extremely fine differences in aggregate morphology,” says Rosenberger. Two distinct forms stood out: crumb-like aggregates with a rough, irregular structure, and ball-like aggregates with a more defined appearance. This raised a key question: what determines which type appears – and are they random or biologically meaningful?
This is where the team made a breakthrough. They successfully reconstructed the sequence of molecular events – the formation of crumbs, balls, and the transitions between them – and identified their temporal order. The crumb-like aggregates appeared first, as an early response of stressed cells. This was associated with activity in special cell compartments called peroxisomes. The ball-like aggregates emerged later, during more advanced stages of fibrosis. Interestingly, however, the type of aggregate did not always correlate with disease severity. Even patients with only mild fibrosis could show the advanced ball-like morphology.
“The shift from crumbs to balls was a key finding,” Rosenberger explains. “It reveals the sequence of compensatory responses liver cells mount in an effort to combat aggregate formation – and with it, liver fibrosis.”
Toward Clinical Applications
The improved AI-based image analysis played a pivotal role in uncovering these mechanisms. “Recent technological advances in mass spectrometry were crucial,” says Professor Matthias Mann. “We can now perform single-cell measurements, allowing us to extract detailed molecular information from just a small amount of tissue – even from individual diseased liver cells.”
The study’s findings may soon have clinical relevance. The development of fibrosis in individuals with the homozygous mutation could potentially be prevented.“By reviewing patient histories, we saw that those with severe fibrosis lacked the early peroxisomal response,” says Rosenberger. “We now know this response is protective. Our goal is to develop an early warning system for liver fibrosis – a way to identify patients at risk before symptoms arise.”
Aleksander Krag, professor at the University of Southern Denmark and head of Odense Liver Research Center, emphasizes: “By capturing alpha-1 antitrypsin accumulation at the single-cell level, we have uncovered early molecular triggers of how alpha-1 antitrypsin deficiency progresses. This points toward actionable targets that could lead to improved therapies for patients.”
Professor Pavel Strnad, a hepatologist at University Hospital Aachen and long-standing collaborator on the project, adds: “Erroneous protein folding is central to many human diseases, including Parkinson’s and Alzheimer’s. Studying a monogenic condition like Alpha-1 Antitrypsin Deficiency provides a unique opportunity to better understand disease progression. This work deepens our insight into protein folding disorders and their consequences, and will be important even beyond Alpha-1 Antitrypsin Deficiency.”
The study was published in the journal Nature.
Glossary
Deep Visual Proteomics: a spatial proteomics method developed in the laboratory of Professor Matthias Mann (Mund et al., Nature Biotechnology, 2022). This method combines modern microscopy, artificial intelligence, laser microdissection and ultra-sensitive mass spectrometry.
Enzymes: are proteins that act as catalysts. This means that their involvement accelerates biological reactions.
Mass spectrometry: is an analytical technique that separates and measures ions according to their mass-to-charge ratio to identify and quantify chemical substances or molecules. It is a cornerstone technology in proteomics, enabling the identification and quantification of thousands of proteins in complex biological samples.
Protease inhibitor: a substance that inhibits the activity of enzymes that cleave proteins, known as proteases, to prevent the degradation of proteins.
Proteome: comprises the totality of all proteins in a living organism, a tissue or a cell at a specific point in time. The proteome is highly dynamic and reacts to the requirements of the cell, as well as to diseases or environmental influences.
Proteomics: is the study of the proteome.













