Novel mechanism for maintenance of bacterial proteostasis identified
Researchers at the Max Planck Institute of Biochemistry (MPIB) have identified a novel mechanism that ensures proteostasis maintenance in E. coli when chaperone availability is limited.
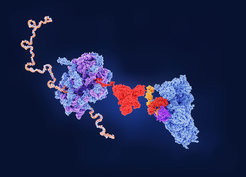
When the chaperone network is defective, RF3 is recruited to the ribosome and mediates the termination of misfolded nascent polypeptide chains.
Investigating the chaperone network
Proteins - the building blocks of life - fulfil a large variety of functions in our body. They are synthesized in the cell at large complexes called ribosomes. During this process, information from the messenger RNA is translated into a sequence of amino acids, which are connected to form a polypeptide chain. For proteins to fulfil their function, these nascent polypeptide chains need to be folded into specific three-dimensional structures. How can cells ensure the correct folding of their proteins? This is where chaperones become relevant. The folding helpers ensure that proteins are folded correctly, repair misfolded proteins and initiate the degradation of faulty proteins at the proteasome.
Using quantitative proteomics, Liang Zhao (postdoctoral fellow in the groups of F.-Ulrich Hartl and Manajit Hayer-Hartl) and the team first established an overview of the chaperone network in the model organism Escherichia coli (E. coli). They analyzed E. coli proteins via mass spectrometry and identified a large variety of chaperones important for protein folding at the co-translational level. They found Trigger factor (TF), DnaJ, and DnaK to be the most abundant chaperones. In case of functional loss of the central hub chaperone DnaK, the chaperones HtpG, GroEL and ClpB contribute increasingly to compensate the loss.
Defects in the chaperone network lead to a response at the ribosome
But what happens at the ribosome when the chaperone network is defective? To investigate this question, the researchers collaborated with Pierre Genevaux and Marie-Pierre Castanié from the University of Toulouse and limited the number of chaperones available in the cell by using knock-out models. They thereby induced misfolding of nascent polypeptide chains. In response, release factor 3 (RF3) was recruited to the ribosome. RF3 subsequently cooperated with another release factor, RF2, leading to the premature termination of protein synthesis and the ensuing release of incomplete misfolded nascent polypeptide chains from the ribosome. That way, the incomplete polypeptide chains could be degraded. Conversely, when this mechanism was inhibited through deletion of RF3, misfolded proteins accumulated in aggregates and impaired the synthesis of new peptide chains. The researchers thus identified, for the first time, a connection between a defect in or limitation of chaperones and an ensuing response involving RF3 at the ribosome. This mechanism is crucial to maintain proteostasis when chaperone availability is restricted as it facilitates clearance of misfolded proteins.
The next steps involve identifying similar mechanism in yeast or mammalian cells. If an analogous mechanism can be found, this might pave the way for future treatments in neurodegenerative diseases such as Alzheimer’s.
Original Publication:
L. Zhao, M.-P. Castanié-Cornet, S. Kumar, P. Genevaux, M. Hayer-Hartl, and F. Ulrich Hartl: Bacterial RF3 Senses Chaperone Function in Co-translational Folding, Molecular Cell, June 2021
DOI: https://doi.org/10.1016/j.molcel.2021.05.016












